Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Detection of Cardiovascular Diseases from ECG Images Using Machine Learning and Deep Learning Methods
Authors: Mrs. M.S.R Pavani, Chilukuri Rahul Chowdary, Neeli Reena, Ummiti Jagadeesh, Pecchetti Krishna Vamsi
DOI Link: https://doi.org/10.22214/ijraset.2024.60790
Certificate: View Certificate
Abstract
Cardiovascular diseases (CVDs) are the leading cause of death globally, emphasizing the importance of early prediction and classification for saving lives. Electrocardiogram (ECG) is a widely used, cost-effective, and noninvasive tool for measuring heart electrical activity, crucial for detecting CVDs. This study leverages deep learning techniques to predict four major cardiac abnormalities—abnormal heartbeat, myocardial infarction, history of myocardial infarction, and normal individuals—using a public dataset of ECG images from cardiac patients. The research explores transfer learning with pretrained deep neural networks such as SqueezeNet and AlexNet, as well as introduces a novel convolutional neural network (CNN) architecture for cardiac abnormality prediction. Additionally, pretrained models and the new CNN architecture are utilized as feature extraction tools for traditional machine learning algorithms including support vector machine, K-nearest neighbors, decision tree, random forest, and Naïve Bayes. Experimental results showcase the superiority of the proposed CNN model over existing works, achieving 98.23% accuracy, 98.22% recall, 98.31% precision, and 98.21% F1 score. Furthermore, when the proposed CNN model is utilized for feature extraction, it achieves the highest score of 99.79% using the Naïve Bayes algorithm.
Introduction
I. INTRODUCTION
Cardiovascular diseases (CVDs) are the leading cause of death globally, claiming an estimated 17.9 million lives each year, which accounts for 32% of all deaths worldwide. Among these deaths, approximately 85% are attributed to heart attacks, known as myocardial infarctions (MI). Early detection of CVDs is crucial for saving lives, and various techniques are employed in the healthcare system for this purpose, including electrocardiogram (ECG), echocardiography (echo), cardiac magnetic resonance imaging, computed tomography, and blood tests. Among these, ECG is particularly common, inexpensive, and noninvasive, making it a key tool for identifying heart-related CVDs. However, manual interpretation of ECG results by skilled clinicians can be prone to inaccuracies and is time-consuming.
There is significant potential for leveraging artificial intelligence (AI) advances in healthcare to reduce medical errors, especially in the automatic prediction of heart diseases using machine learning and deep learning techniques. Machine learning methods typically require expert intervention for feature extraction and selection to identify relevant features before the classification phase. Feature extraction involves reducing the number of features in a dataset by transforming or projecting the data into a new, lower-dimensional feature space that retains the essential information of the input data.
This process aims to create a new set of features that combination of the original features, extracting most, if not all, of the information present in the input data. Principal component analysis (PCA) is one of the most well-known feature extraction methods. Feature selection, on the other hand, involves removing irrelevant and redundant features (dimensions) from the dataset during the training process of machine learning algorithms.
Various methods exist for feature selection, categorized as unsupervised (which does not require the output label for feature selection) and supervised (which uses the output label for feature selection). Under supervised feature selection, three methods are commonly used: the filter method, the wrapper method, and the embedded method.
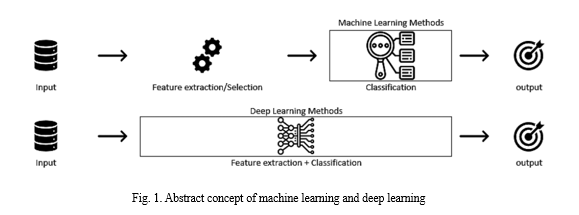
II. LITERATURE REVIEW
Many research works have been conducted for automatically predicting cardiovascular diseases using machine learning and deep learning methods by utilizing ECG as digitals or images data representation. Bharti et al compared machine learning and deep learning methods on the UCI heart disease dataset to predict two classes. The deep learning method achieved the highest accuracy rate of 94.2%. In their architecture of deep learning model, they used three fully connected layers: the first layer consists of 128 neurons followed by a dropout layer with 0.2 rate, the second layer consists of 64 neurons followed by a dropout layer with 0.1 rate, and the third layer consists of 32 neurons. The machine learning methods with features selection and outliers’ detection achieved accuracy rates as: RF is 80.3%, LR is 83.31%, K-NN is 84.86%, SVM is 83.29%, DT is 82.33%, and XGBoost is 71.4%. The research in [29] concluded that deep learning has proven to be a more accurate and effective technology for a variety of medical problems such as prediction. Deep learning methods will replace the traditional machine learning based on feature engineering. Kiranyaz et al. [30] proposed a CNN that consisted of three layers of an adaptive implementation of one-dimensional (1-D) convolution layers. This network was trained on the MIT-BIH arrhythmia dataset to classify long ECG data stream. They achieved accuracy rates of 99% and 97.6% in classifying ventricular ectopic beats and supraventricular ectopic beats, respectively. Also, the work in [31] proposed a CNN that consisted of three 1-D convolution layers, three max-pooling layers, one fully connected layer, and one softmax layer. The filter size for the first and second convolutional layers was set to 5 and a stride of 2 was used for the first two max-pooling layers. They achieved an accuracy rate of 92.7% in classifying ECG heartbeats using the MIT-BIH arrhythmia dataset Khan et al. [22] applied transfer learning approach using the pretrained single shot detector (SSD)-MobileNet-v2 [32] to detect cardiovascular diseases from the ECG images dataset of cardiac patients by predicting the four major heart abnormalities: abnormal heartbeat (AH), MI, history of MI (H. MI), and normal person (NP) classes. As preprocessing steps, the data size was adjusted and the 12 leads of each ECG image were labeled. SSD is used to classify and localize the objects in one step. The dataset was split 80% for training and 20% for testing. They used a batch size of 24, 200K training iterations for the training step, and a learning rate of 0.0002 to train their model. Their training phase lasted almost 4 days. They achieved a high precision rate for the MI class, i.e., 98.3%.
III. METHODS
- CNN- In deep learning, Convolutional Neural Networks (CNNs) are a specialized type of artificial neural network designed for image classification and processing. CNNs organize neurons in three dimensions: height, width, and depth (channel). For instance, an input image might be represented as 227 × 227 × 3, indicating that its width and height are both 227 pixels, and it has 3 channels for red, green, and blue. The primary role of CNNs is to extract essential features from input images. CNNs consist of two main components: convolutional layers and pooling layers. Convolutional layers apply convolution operations on the input data using a filter or kernel to create a feature map that represents the detected features of the input. During convolution, the filter slides over the input, and at each position, matrix multiplication is performed, with the results summed onto the feature map. Pooling layers, on the other hand, reduce the spatial dimensions of the feature map, reducing the computational complexity of the network.
Common pooling operations include max pooling and average pooling, which downs sample the feature map by taking the maximum or average value in each pooling region, respectively. Higher layers in CNNs typically consist of fully connected layers, and the final layer often uses a sigmoid or softmax activation function to produce the predicted output. The example in Figure 2 illustrates a simple convolution process for an input with a depth of 1. The convolution process is linear. To add nonlinearity to the output, the convolution layer is followed by an activation function layer such as ReLU or its variants. After the convolution layer, a pooling layer such as max-pooling layer could be used to down sample the feature map to reduce the computational cost. Fig. 3 shows a simple example of max-pooling for an input with depth of 1.
2. Pretrained Deep Learning Models- The pretrained deep NNs can be used for transfer learning, feature extraction, and classification. In this article, low-scaled SqueezeNet and AlexNet pretrained CNN networks that can be executed on a single CPU are used for transfer learning and feature extraction. The transfer learning approach is commonly used with pretrained deep NNs applied to a new dataset. Therefore, it could benefit from the pretrained network that has already learned a variety of features that can be transferred to other similar tasks. Most of the pretrained networks have been trained with more than a million images and can classify images into 1000 object classes. In applying the transfer learning approach, the final layers of the pretrained network are replaced with new layers to learn the specific features of the new dataset. Then, the model is fine-tuned by training it on a new training dataset with specific training parameters and testing its performance measure on a new test dataset.
3. Proposed CNN Architecture- The proposed CNN model contains besides the input and output layers, six 2-D convolutional layers, three fully connected layers, three max-pooling layers, eight leaky ReLU layers, eight batch normalization layers, five dropout layers, two depth concatenation layers, and one softmax layer. In total, there are 38 layers. The architecture of the proposed model is shown in Fig. 4. The proposed CNN model consists of two branches that help extract more representative features, namely the stack branch and the full branch. The proposed CNN model accepts input image of size 227×227×3. The input image flows into the two branches simultaneously.
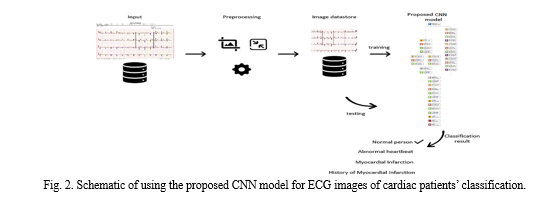
IV. RESULTS AND DISCUSSIONS
For performance analysis, accuracy, precision, recall, F1 score, and training and testing times were used. These measurements are based on the analysis of the data in a confusion matrix. Where the accuracy is the percentage of positively predicted observations relative to the total number of observations. Recall represents the ratio of positively predicted observations to all observations in the true class (should be positively estimated). Precision expresses the ratio of positively predicted observations to all observations in the predicted class (should be positively predicted). The F1 score is the weighted average of both Recall and Precision. Thus, it takes into account both the false negatives and the false positives values.
- Results of Transfer Learning and Proposed CNN Model- The state-of-the-art architectures of the pretrained networks SqueezeNet and AlexNet were used to apply the transfer learning approach in our study. Both were originally trained for the classification of 1000 image classes. To retrain these networks for classifying the new set of ECG images in the dataset, we replace the last layers of these models to make them suitable for the new task. In AlextNet, the last fully connected layer is replaced with a new fully connected layer containing the same number of neurons as the number of our predicted classes, i.e., 4. However, since SqueezeNet does not use fully connected layers, we replace the last convolutional layer which is used to identify 1000 classes with a new convolutional layer containing 4 1×1 filters.
For both pretrained networks used, a new classification layer is added in place of the existing one, which produces an output based on the probabilities computed by the softmax layer.
The average accuracy rate for the proposed CNN model shows similar high results when the RL values are changed. In contrast, the pretrained SqueezeNet and AlexNet models show poor results when the initial learning rate were 0.01 and 0.001, but they start to show slightly good results when the LR is set to 0.0001. This is because, in transfer learning, the weights of the pretrained models are not learned from scratch. Therefore, to avoid getting stuck in local minima, it is better to start with a lower LR such as 0.0001 when applying transfer learning techniques. The average accuracy rates are 96.79% and 95.43% for AlexNet and SqueezeNet, respectively with RL of 0.0001. On the other hand, the proposed CNN model also outperforms the other models in terms of time cost, as can be seen in Table VI. Although SqueezeNet has the smallest number of parameters and is a fully CNN, it achieves the worst performance in terms of time cost. This is because the number of computations in the convolutional layers is very high, so it takes more time to be processed, especially when running on a single CPU platform. Fig. 9 depicts the training progress of our proposed CNN model on the ECG images dataset in fold-1 (LR = 0.0001). The accuracy rate increases gradually with each successive iteration. Moreover, the loss decreases smoothly as the iteration progresses and reaches 0.0043.
2. Results of Using Pretrained Deep Learning Models As a Feature Extractor- The pretrained SqueezeNet and AlexNet networks were used to extract the features of the ECG images in the dataset. As well as, our proposed CNN model was used as a feature extractor and the results were compared. The power of deep learning can be used to extract image features without re-training the entire network. The activations of the network are computed by forward propagation of the input images up to the specific feature layer. The activation feature layers used are conv10 (layer number 64), fc7 (layer number 20) and fc02 (layer number 32) for SqueezeNet, AlexNet and our proposed CNN model, respectively. The performance measures are calculated and presented in Table X. As can be seen, the most successful result was obtained with a rate of 99.79% for the accuracy, recall, precision, and F1-score of the NB algorithm when our proposed CNN model was used as the feature extractor. The accuracy rates of 99.47%, 97.87%, and 97.66% were obtained by the SVM algorithm when our proposed CNN model, SqueezeNet, and AlextNet, respectively, were used to extract the features. As a result, the best achievements for all performance measures were obtained when using our proposed CNN model as the feature extractor. When comparing SqueezeNet and AlexNet, we almost achieved better accuracy rates for SVM, RF and NB algorithms using the features extracted from SqueezeNet than those from AlexNet.
Conclusion
This study introduces a lightweight CNN-based model for classifying four major cardiac abnormalities—abnormal heartbeat (AH), myocardial infarction (MI), history of myocardial infarction (H. MI), and normal person (NP) classes—using a public dataset of ECG images from cardiac patients. Experimental results demonstrate the effectiveness of the proposed CNN model in cardiovascular disease classification and its potential as a feature extraction tool for traditional machine learning classifiers. This suggests that the proposed CNN model could serve as an assistive tool for clinicians in the medical field, offering a more efficient alternative to the manual process, which is prone to inaccuracies and time constraints.Future research directions could include the use of optimization techniques to fine-tune the hyperparameters of the proposed CNN model for optimal performance. Additionally, the proposed model could be applied to predict other types of problems. Given that the proposed model is a low-scale deep learning method in terms of the number of layers, parameters, and depth, it could be explored for classification purposes in the Industrial Internet of Things domain. One of the key strengths of the proposed model lies in its dual functionality. Not only does it excel in directly classifying cardiac abnormalities, but it also demonstrates utility as a feature extraction tool for traditional machine learning classifiers. This versatility suggests that the model could play a significant role in enhancing the efficiency and accuracy of cardiovascular disease diagnosis, potentially reducing the reliance on manual interpretation methods that are prone to error and time-intensive.
References
[1] World Health Organization (WHO), “Cardiovascular diseases,” Jun. 11, 2021. Accessed: Dec. 27, 2021. [Online]. Available: https://www.who.int/health-topics/cardiovascular-diseases [2] Government of Western Australia, Department of Health, “Common medical tests to diagnose heart conditions,” Accessed: Dec. 29, 2021. [Online]. Available: https://www.healthywa.wa.gov.au/Articles/A_E/Common-medical-tests-to-diagnose-heart-conditions [3] M. Swathy and K. Saruladha, “A comparative study of classification and prediction of cardio-vascular diseases (CVD) using machine learning and deep learning techniques,” ICT Exp., to be published, 2021. [Online]. Available: https://doi.org/10.1016/j.icte.2021.08.021 [4] R. R. Lopes et al., “Improving electrocardiogram-based detection of rare genetic heart disease using transfer learning: An application to phospholamban p.Arg14del mutation carriers,” Comput. Biol. Med., vol. 131, 2021, Art. no. 104262. [Online]. Available: https://doi.org/10.1016/j.compbiomed.2021.104262 [5] R. J. Martis, U. R. Acharya, and H. Adeli, “Current methods in electrocardiogram characterization,” Comput. Biol. Med., vol. 48, pp. 133–149, 2014. [Online]. Available: https://doi.org/10.1016/j.compbiomed.2014.02.012 [6] M. Kantardzic, Data Mining: Concepts, Models, Methods, and Algorithms, 3rd ed. Hoboken, NJ, USA: Wiley, 2020. [7] S. García, J. Luengo, and F. Herrera, Data Preprocessing in Data Mining, 1st ed. Berlin, Germany: Springer, 2015. [8] G. Dougherty, Pattern Recognition and Classification: An Introduction. Berlin, Germany: Springer, 2013. [9] A. Subasi, Practical Machine Learning for Data Analysis Using Python. Cambridge, MA, USA: Academic, 2020. [10] J. Soni, U. Ansari, D. Sharma, and S. Soni, “Predictive data mining for medical diagnosis: An overview of heart disease prediction,” Int. J. Comput. Appl., vol. 17, no. 8, pp. 43–48, 2011.
Copyright
Copyright © 2024 Mrs. M.S.R Pavani, Chilukuri Rahul Chowdary, Neeli Reena, Ummiti Jagadeesh, Pecchetti Krishna Vamsi. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET60790
Publish Date : 2024-04-22
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

